This summer, in Dr. Pollock’s research lab, I will be implementing a new technique that allows the incorporation of an unnatural amino acid into a protein. Aminoacids are the building blocks of proteins, and the sequence of these aminoacids determines the structure and function of any protein. Naturally, proteins are composed of 20 aminoacids that have specific side-chains and properties. Instructions to make proteins are stored in the DNA of the cells and are followed by two very important processes that I will explain in more detail. Since the 20 naturally occurring aminoacids are limited, scientists have developed ways to expand the genetic code to include what we refer to as unnatural aminoacids (UAAs). These UAAs contain a diversity of side-chains, which allow scientists to probe the roles of proteins in disease progression or manipulate their natural functions to study their therapeutic potential.
Efficient protein production in the laboratory requires help from our bacterial friends. The bacteria of E. coli has been used by scientists for decades as the workhouse for protein production, due to its simplicity which makes it a good candidate for genetic modifications. It is common to use E.coli in molecular biology to study a variety of proteins and their functions in our organism, by making use of its cell machinery.
The genomic DNA of bacterial cells is circular, so in order to get them to make our protein of interest, we give them a new circular loop of DNA called a plasmid. This plasmid provides all the information the bacteria need to carry out the job of making our protein of interest. In lab, this is done by utilizing a method called bacterial transformation. This process is a key step in molecular cloning and involves the insertion of DNA of interest, usually in the form of a plasmid, into competent cells. Competent cells are simply cells whose membranes have been slightly disrupted so that they can take up plasmids either by heat shock or electric shock. After it is inserted, the plasmid is amplified through DNA replication, simply meaning that more copies of the DNA are made using machinery already present in the bacteria. The last step in this process is protein expression through transcription and translation.
To make our desired protein, the cell has to complete two very important processes: transcription and translation. As I mentioned above, the information to make proteins is stored in the DNA. During transcription, the DNA strand is used to make an RNA strand, which contains the same information as the DNA but in a slightly different language. The RNA strand can be deciphered by a complex called the ribosome. During the process of translation, the ribosome acts as an assembly line to travel along the RNA strand and read a code that matches the sequence with aminoacids. The aminoacids come to the assembly line on carriers called aminoacylated tRNAs, who are given the correct aminoacid to add by tRNA synthetase. Three codons in the RNA do not code for an aminoacid, rather they act as stop signs and recruit these small proteins called release factors. Release factors act as police officers to stop translation and snatch the aminoacid chain off the ribosome so it can be released as our desired protein.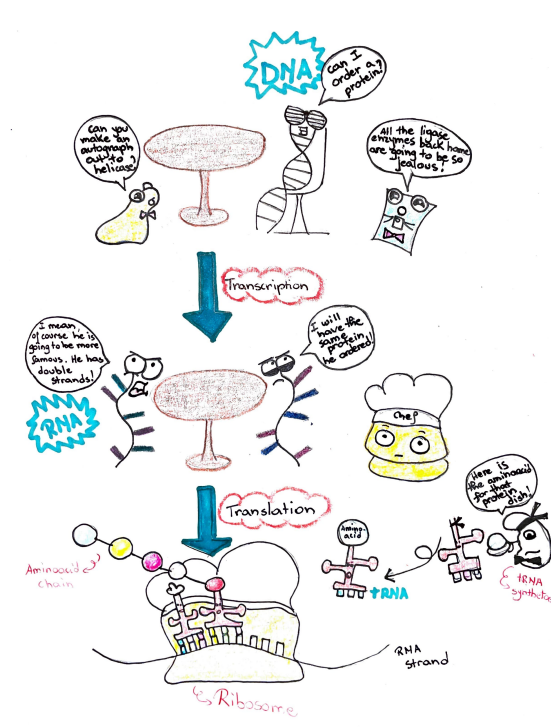
Now that we are familiar with the process of protein expression, we can better understand what is different when incorporating something “unnatural” to a protein. The unnatural aminoacid is simply a camouflage for one of the natural aminoacids we choose, meaning it looks very similar to the natural aminoacid, but has this superpower to do something different like form interactions with other proteins once subjected to light or tag the protein as a highlighter. Before, I mentioned that there are three codes in the RNA sequence which spell “STOP” and terminate translation. E. coli cells do not use all three of these stop codes equally, they kind of have a favorite one to use for terminating translation. One of the least used stop codes is the amber codon. Therefore, scientists have used this code as a place to introduce an UAA. In addition to changing the DNA and RNA code, a tRNA and its related tRNA synthetase that recognize the unnatural aminoacid needed. They are generated through mutations in an accelerated evolutionary process. When the appropriate tRNA, tRNA synthetase, UAA, and appropriate code in the DNA are all present in the E. coli, they will use their transcription and translation machinery to express the protein containing the UAA of choice.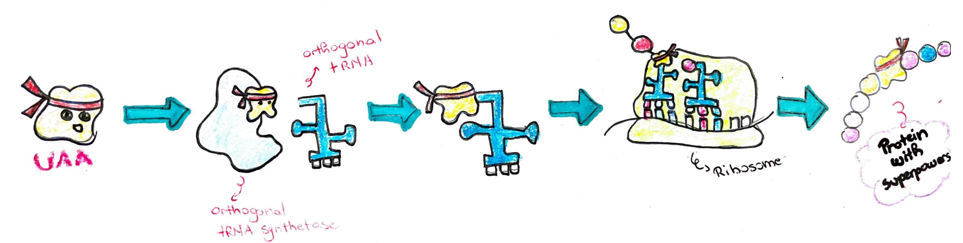
Now that we have broadened our understanding of unnatural amino acid incorporation, we can acknowledge the importance such a powerful tool holds. In the past years, the use of unnatural aminoacids has allowed scientists to discover very interesting things. In Thompson’s (2014) paper, the researchers were able to identify several proteins that bind PRMT1, a protein whose activity is associated with the progression of many diseases, such as cancer, heart disease, etc. Therefore, UAAs enable us to modify the properties of proteins at will and allows us to carry out all these cool experiments that may shine a light on figuring out many important functions of proteins within our cells.
– Sonia Mecorapaj
Citations:
Nguyen TA, Cigler M, Lang K. Expanding the Genetic Code to Study Protein-Protein Interactions. Angewandte Chemie (International ed. in English). 2018 Oct;57(44):14350-14361. DOI: 10.1002/anie.201805869. PMID: 30144241.
Rust, H. L., Subramanian, V., West, G. M., Young, D. D., Schultz, P. G., & Thompson, P. R. (2014). Using unnatural amino acid mutagenesis to probe the regulation of PRMT1. ACS chemical biology, 9(3), 649–655. https://doi.org/10.1021/cb400859z
Wals, K., & Ovaa, H. (2014). Unnatural amino acid incorporation in E. coli: current and future applications in the design of therapeutic proteins. Frontiers in chemistry, 2, 15. https://doi.org/10.3389/fchem.2014.00015

Recent Comments